Data Institute: Clinical Neuroinformatics & AI Laboratory
Projects & Research
Early Predictions of Hospital Acquired Sepsis
Zachary Barnes, MS Data Science
Collaboration with Dr. Leo Liu @ UCSF
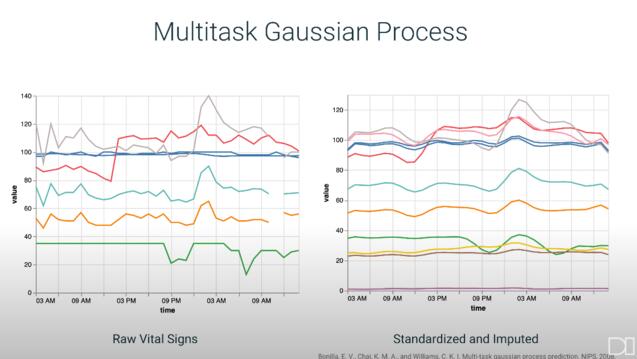
In collaboration with the UCSF School of Medicine, our research is on the development of a model for the early prediction of hospital acquired sepsis. Several features differentiate this research from current sepsis models in research and production: training a model using the UCSF information commons database, extensive feature engineering, labeling patients based on the most reliable definition of sepsis, and focusing on a non-ICU ward. We achieved state of the art results with an AUCPR of 0.67 four hours before onset using the leading model, a multi-task gaussian process and recurrent neural network, and comparable results with tree-based models trained with expert engineered features.
De-Crappifying Microscopy Images of the Brain
Annette Lin and Alaa A Latif, MS Data Science
Collaboration with Dr. Uri Manor and Linjing FanG @ Salk
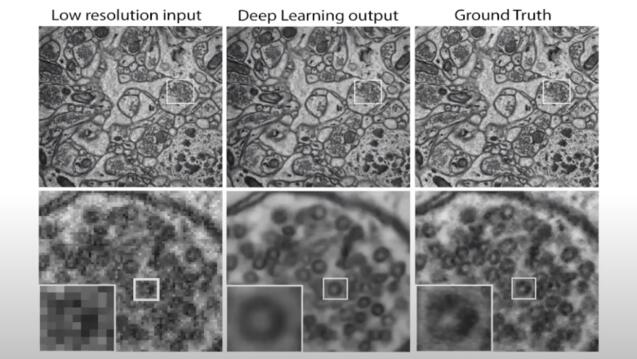
Electron Microscopy (EM) is an important tool for studying biological systems at their most fundamental level. Our research work involves training deep neural networks to de-crapify (de-noising and super-resolution) EM images of brain structures using self-supervised learning and perceptual losses.
Multivariate Pattern Analysis with Machine Learning
Maxwell Calehuff, MS Data Science
Collaboration with Dr. Ross Filice @ Medstar Georgetown University Hospital
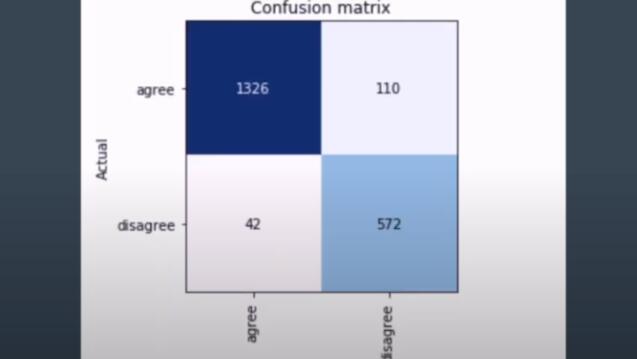
This project applies cutting edge natural language processing techniques to improve feedback for medical students. Students' written imaging reports are automatically flagged when they deviate from those of the supervising physicians. Portions of the flagged reports that deviate in meaning, not just phrasing, are highlighted to improve understanding.
Fast Acquisition Microscopy via Deep Learning
Dr. Tahir Issa Bachar, USF DI Postdoctoral Associate
Collaboration with Dr. Claudio Vinegoni @ Harvard
Laser scanning microscopy is a powerful imaging modality ideal for monitoring spatial and temporal dynamics in both in vitro and in vivo models with particular emphasis in the neuroimaging field. Unfortunately, the video-rate acquisition required to capture these changes lead to low resolution images, with high spatial distortion and low signal-to-noise ratio. By combining microscopy fast acquisition methods with deep learning, we show here that image acquisition up to 20x the speed of equivalent standard high resolution condition can be obtained without sacrificing image quality. Specifically, we present here a GAN based training approach able to simultaneously:
- Super-resolve
- Denoise
- Correct distortion on fast scanning acquisition microscopy images
Contrast Enhancement Prediction in Brain MRI
Wendeng Hu and Todd Zhang, MS Data Science
Collaboration with Dr. Greg Zaharchuk @ Stanford
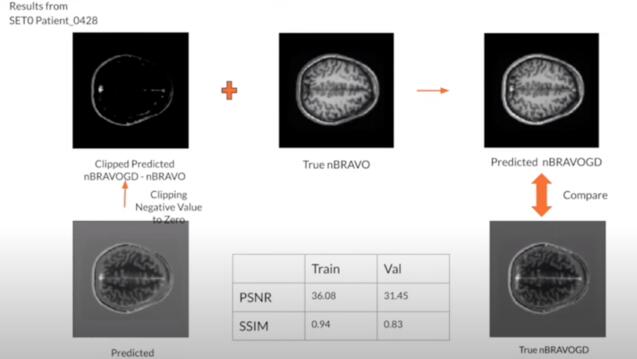
Gadolinium-based contrast agents (GBCAs) have become an integral part of daily clinical brain abnormality diagnosis. However, the consensus is that the use of GBCAs could have harmful effects on the human body and should only be used in limited amounts when no alternatives are available. The team developed a deep learning model to predict contrast enhancement from non-contrasted multiparametric brain magnetic resonance imaging (MRI) to replace the use of GBCAs potentially. The predictions highly resembled the actual contrast-enhanced images based on PSNR and SSIM metrics.
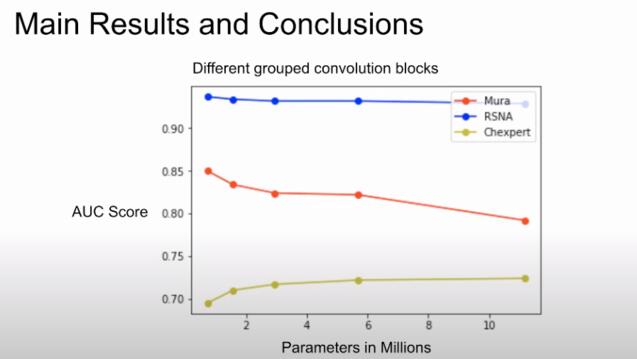
Simple CNN Models for Classification on Medical Images
Roja Immanni, MS Data Science
Collaboration with Dr. Gilmer Valdes, UCSF
Medical image datasets are fundamentally different from natural image datasets in terms of the number of available training observations and the number of classes for the classification task. We hypothesized that compared to architectures used for natural images, those needed for medical imaging can be simpler. Here, we propose smaller architectures and show how they perform similarly while significantly saving training time and memory.
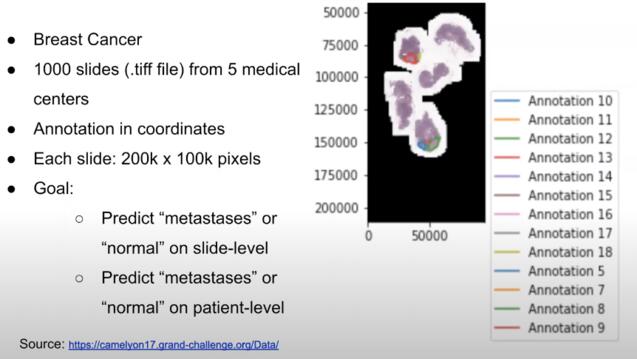
Multiple-Instance Learning on Pathology Slides
Lori Sheng, MS Data Science
Collaboration with Dr. Dmytro Lituiev @ UCSF
When it comes to applying computer vision in the medical field, most tasks involve either 1) image classification for diagnosis or 2) segmentation to identify and separate lesions. However, in pathology cancer detection, this is not always possible. Obtaining segmentation labels is time-consuming and labor-intensive. Furthermore, a single pathology slide can be up to 200k x 100k pixels resolution. We investigate Multiple Instance Learning as a deep learning method to handle both memory and labeling constraints.
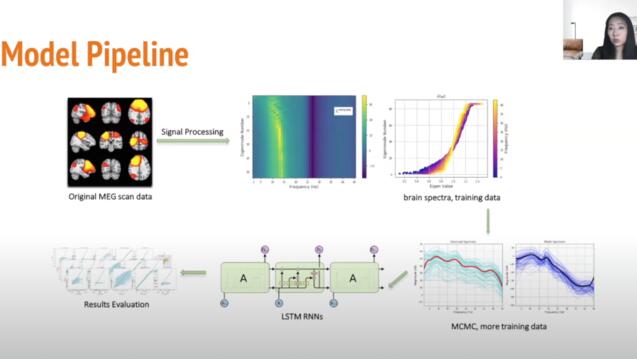
Disease Diagnosis from Brain MEG with LSTMs
Lexie Sun, MS Data Science
Collaboration with Dr. Ashish Raj and Xihe Xie @ Brain Networks Lab - UCSF
When it comes to applying computer vision in the medical field, most tasks involve either 1) image classification for diagnosis or 2) segmentation to identify and separate lesions. However, in pathology cancer detection, this is not always possible. Obtaining segmentation labels is time-consuming and labor-intensive. Furthermore, a single pathology slide can be up to 200k x 100k pixels resolution. We investigate Multiple Instance Learning as a deep learning method to handle both memory and labeling constraints.
Auto-Segmentation of Brain Metastases
Akanksha, MS Data Science
Collaboration with Dr. Ashish Raj and Xihe Xie @ Brain Networks Lab - UCSF
The task is to build a deep learning model to perform 3D semantic image segmentation of brain sMRI into White Matter, Grey Matter, and Cerebrospinal fluid, and the employing transfer learning to extend the segmentation model for the regression task of predicting cognitive scores of the patients. Presently I have built a 3D symmetrical U-Net model using PyTorch which gives an overall dice coefficient of 92% across classes on the ADNI dataset, and right now I am in the process of extending it to the regression task.
Functional Embeddings Identify Differences in Schizophrenia Sub-types
Lead: Dr. James Wilson, USF
Speaker: Dr. Abbie Popa, USFDI PostDOC
The team helped create a new technique to quantify how different areas of the brain are connected and then utilized this technique to examine differences between schizophrenics and healthy individuals. Using a mathematical representation of how correlated two parts of the brain are across individuals in a group, the team was able to show correlations among brain regions exactly where they were expected. With the model validated she then compared healthy individuals to those with schizophrenia. The results were able to show a difference in regions of the brain that are well documented in neuroscience literature as areas the disease effects. Future work includes being able to identify sub-types of schizophrenia using this technique, as well as collecting more data to increase the effectiveness of the technique.
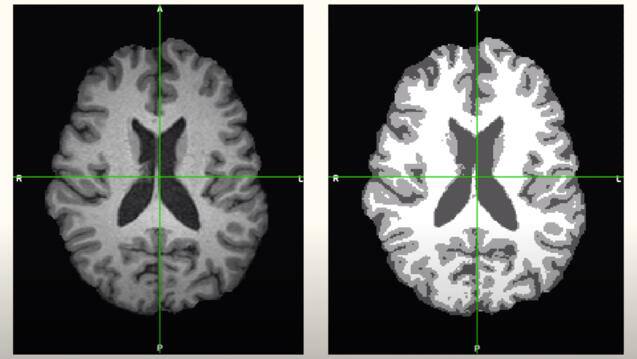
Automated Segmentation of Brain Ventricles using 3D U-Nets
Lead: Dr. Olivier Morin, UCSF
Speakers: Anish Dalal and Robert Sandor, MS Data Science
The team developed a deep learning model that performs as well or better than the leading proprietary software used in hospitals to identify Hydrocephalus (cerebrospinal fluid leaking into the ventricles of the brain). Monitoring of the ventricles in the brain for swelling is very important for newborns, patients after brain surgery, and for patients who have suffered a stroke or other traumatic brain injury. If fluid builds up in these ventricles it can lead to symptoms including headache, loss of vision, a decline in memory, and impaired neural development . Monitoring the ventricles is an expensive and time-consuming task for doctors because they have to go through many 2D images of the brain, drawing boundaries around the ventricles. This model makes this process faster and more accurate than the current software solution, which the team hopes will help improve monitoring of patients and bring down the cost of care for hospitals.
Deep Learning to Assist Medical Imaging Diagnostics
Lead: Dr. Vasant Kearney, UCSF
Speakers: Tianqi Wang and Alan Perry, MS Data Science
The team studied the applications of deep learning on CT scans, while also producing two academic papers on their findings. The first paper showed that deep learning can categorize pixels of CT scans based on if they are a part of the different organs in the body. This technique could be helpful for clinicians interested in studying any disease that manifests in organ tissue. The second paper showed that MRI data could be converted to CT scan data. Using deep learning techniques, the team showed that a model could be used to convert data between the two types of scans. While this has applications for clinicians who may want one type of scan to compare two patients without having to run another test, this technique also has the possibility of affecting data science by allowing practitioners to increase the amount of data they have for a specific task.
Improving the Efficiency in View Classification of Echo-Cardiograms with Deep Learning
Lead: Dr. Rima Arnaout, UCSF
Speakers: Divya Shargavi and Max Alfaro, MS Data Science
Echo-cardiograms are an important imaging technique used by doctors to inform their diagnostics of a patient's heart. However these images contain a lot of variation since echo-cardiograms can be still-images or videos taken from many different angles on moving patient. Doctors have standardized 15 “views” of these images, or angles the ultrasound is taken from. The team worked to train a model to recognize which of the 15 different “views” the image was taken from. This is important to understand the diagnostic potential of each image, and could also be used to help doctors standardize the variation within the different views. By doing this they hope to ultimately build a tool to assist doctors in diagnostics.
Deep Learning with Medical Images: Towards Best Practices for Small Data
Lead: Dr. Gilmer Valdes, UCSF
Speaker: Miguel Romero, MS Data Science
Large data-sets are getting all the attention from big companies these days, but most medical researchers and practitioners around the world are working with small amounts of data. This poses a challenge to building accurate and reproducible machine learning models. The team took two approaches to address this issue. The first approach was to rigorously assess the efficacy of different methods to train machine learning models. The next approach was to understand how well models transfer knowledge from task to task, based on both the size and contents of the training set.
Multivariate Pattern Analysis with Machine Learning
Lead: Dr. Peter Wais, UCSF
Speaker: Jenny Kong, MS Data Science
Using complex and noisy data from fMRI scans, the team conducted research to correlate the activation of different regions of the brain with High Fidelity Memory (HFM) storage. HFM is a type of active memory, where individuals detect and remember minute details in images. Researchers hope that increasing our knowledge of how this type of memory forms will aid in our understanding of attention’s impact on memory. Using clustering and statistical analyses, the team sought to identify when individuals were using HFM by analyzing the haemodynamic response function from fMRI scans taken while participants took a visual memory test.